Coronaviruses and astroviruses are viruses whose genome is a
single-stranded mRNA, complete with a 3'-UTR and poly-A tail. In a
subset of coronaviruses that include SARS-CoV-2 (COVID-19),
SARS-CoV-1, and MERS-CoV, the 3'-UTR contains a highly-conserved
sequence (in an otherwise rather variable message) that folds into
a unique structure, called the s2m (stem-loop two motif). The s2m
is extremely conserved, and is believed to mediate iteractions
between the 5'-UTR and 3'-UTR in viral replication and
transcription. SARS-CoV-2 posesses almost exactly the same s2m
sequence (and therefore structure) as found in the original SARS
virus genomic RNA. In 2004, we solved the structure of the SARS-CoV-1
s2m RNA. (PDB
entry 1XJR). Because the two viruses are nearly identical in
this region, we also have a good idea for the structure of the
SARS-CoV-2 s2m RNA. The s2m structure reveals several unique
features that include potential sites for antiviral drugs to bind.
This gives us (or anyone) the opportunity to design antivirals for
an immediate response to the 2020 pandemic. |
The virus that causes COVID-19 is a very large single-strand of an
RNA genomic message. This RNA message is translated into virus
proteins, including possible targets for proposed anti-viral drugs.
Our approach is complementary, in that it directly and efficiently
targets the RNA itself for cleavage and immediate destruction of
the virus. There are at least two ways to do this. One is to use
the cell's own RNA interference pathway to target the virus with
siRNAs. Unfortunately, coronaviruses have evolved a very effective
mechanism to suppress this RNAi pathway during infection. A second
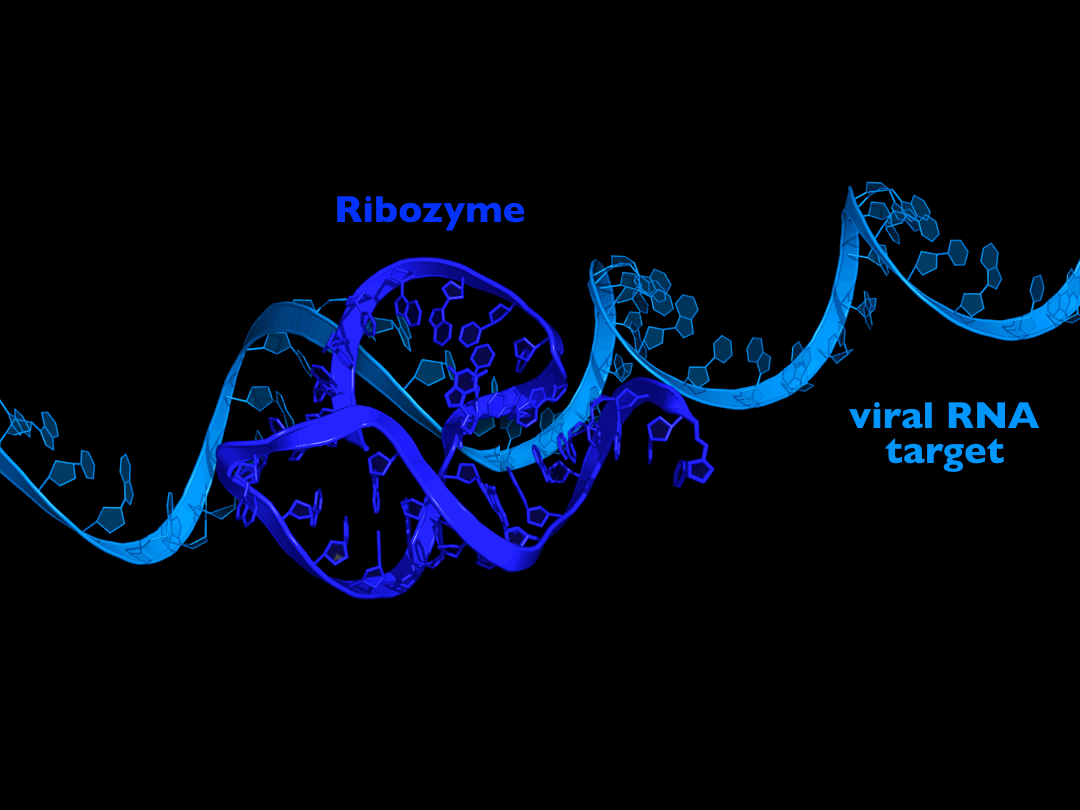
approach is to target the viral RNA for cleavage with small RNA
sequences, called ribozymes, that have the ability to cut the viral
RNA to kill the virus without needing the the RNAi pathway. We have
developed a class of extremely active ribozymes to do exactly this,
and have designed them to target the most vulnerable regions of the
virus RNA. We are currently testing and optimizing these under
laboratory conditions, with the hope of soon identifying the most
promising antiviral drug candidates. |