Recent Highlights
Research Overview
Publications
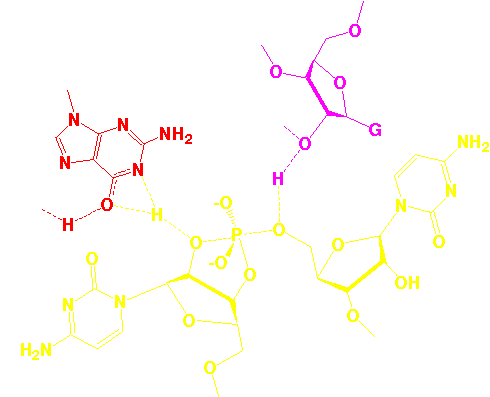
Atomic Coordinates
Research Projects:
Lab members and their pages
Software
Links of Interest
Back to Scott
Lab Home Page
|
|
Since the discovery that RNA
can be an enzyme, a fundamental question has emerged: How does an
RNA molecule fold up into a precise three-dimensional structure
capable of catalyzing a chemical reaction? This problem is
interesting not only from the point of view of living organisms,
but also in terms of trying to understand how a pre-biotic "RNA
World" populated by ribozymes, as evolutionary precursors of
today's protein enzymes found in all living organisms, might have
functioned. The first ribozyme structure to be elucidated was that
of the hammerhead RNA, a small self-cleaving RNA. Currently we are
attempting to understand the catalytic mechanism of this RNA via
static and time-resolved crystallography, and to elucidate the
structures of several other catalytic RNAs. In 1998, we accidently
discovered that the hammerhead, hairpin and HDV ribozymes do not
require divalent metal ions for catalysis. Our
latest structural
results suggest how this is possible in the case of the hammerhead
ribozyme, wherein invariant residues appear positioned for general
base and general acid catalysis. We are also interested in what
aspects of catalysis are so fundamental that they might be
considered univeral principles of macromolecular enzymology (both
for proteins and RNAS).
|